01 August 2025
Reading time [minutes]: 12
Biotech Innovation and Diagnostics
Decentralised Real-Time PCR for respiratory diseases
Rapid and multiplex diagnosis of influenza, RSV and SARS CoV 2 directly at the point of care
Abstract
The adoption of decentralised multiplex real-time PCR platforms represents a paradigm shift in the diagnosis of acute respiratory infections. These solutions, often consisting of portable devices connected to cloud systems, allow simultaneous identification of influenza A/B, respiratory syncytial virus (RSV), SARS-CoV-2 and other pathogens (including bacterial co-infection markers) directly at the point-of-care. This drastically reduces diagnostic Turn-Around Time (TAT) - from days to hours - and improves clinical management: clinicians can isolate infectious patients immediately, initiate targeted antiviral therapies within hours of admission and avoid unnecessary antibiotics in viral cases[1][2]. All in all, decentralised real-time PCR combines the high sensitivity of laboratory tests with the convenience of point-of-care, offering a powerful tool to strengthen antibiotic stewardship and make critical decisions in record time, both in hospital and outpatient settings. Below, we examine the technological structure of these solutions, the diseases covered, the limitations of traditional antigenic approaches and the concrete benefits brought by platforms such as bCUBE™ and bAPP™. An in-depth discussion is devoted to use cases and clinical impact, with direct testimony from Ulisse Biomed's CTO, Lorenzo Colombo.
- Snapshot
- Introduction
- 1. Technological structure and decentralised model
- 2. Diseases covered and detectable co-infections
- 3. Limitations of rapid antigenic approaches
- 4. The added value of multiplex real-time PCR
- 5. bCUBE™ and bAPP™: a case of Italian innovation
- 6. Use cases on the ward and in the territory
- 7. Clinical and operational impact
- Conclusions
Snapshot
- Ct (Cycle Threshold): quantitative parameter of real-time PCR. It corresponds to the number of cycles required for the amplification signal to exceed the threshold level; lower Ct values indicate higher viral loads in the sample.
- TAT (Turn-Around Time): total cycle time between taking the sample and reporting the result. In traditional laboratory diagnostics, the TAT for a respiratory molecular test can be many hours; with decentralised solutions it can be under 1 hour.
- qPCR (Quantitative real-time PCR): Polymerase Chain Reaction technique that allows real-time monitoring of the amplification of genetic material, using fluorescent probes. It is defined as 'quantitative' because it also allows the initial amount of target (virus, bacterium) in the sample to be estimated, thanks to the Ct value.
- Multiplex (PCR multiplex): approach involving the simultaneous amplification of several molecular targets in a single PCR reaction. It is achieved by using different primer/probe pairs labelled with different fluorophores. In the context of respiratory testing, multiplex enables the detection of several viruses (e.g. influenza, RSV, SARS-CoV-2) in parallel from the same sample.
- Co-infection: simultaneous presence of more than one infectious agent in the same patient. In the case of respiratory infections, this may be viral (e.g. influenza + RSV) or virus-bacteria co-infection (e.g. influenza predisposing to pneumococcal pneumonia). Co-infections may aggravate the clinical picture and require combination therapies; identifying them is important for optimal management.
Introduction
In the context of acute respiratory infections - from seasonal influenza to viral pneumonias such as COVID-19 - speed and accuracy of diagnosis play a key role. Recognising the pathogen early on means that infectious patients can be isolated, specific treatments (e.g. antivirals for influenza) can be started early, and unnecessary therapies can be avoided. Unfortunately, traditional laboratory diagnosis methods (central PCR or cultures) often take several hours or days to provide a result, delaying crucial clinical decisions[1]. During the COVID-19 pandemic, it became evident that a centralised-only approach to diagnostics can create logistical bottlenecks and unsustainable waiting times. This has accelerated the development of molecular point-of-care tests: portable instruments capable of performing molecular biology analyses directly at the point of care (outpatient clinic, emergency room, ward) with performance comparable to the laboratory. In particular, decentralised Real-Time PCR platforms in multiplex mode are emerging as a game-changer in respiratory diagnostics. These solutions combine the gold standard PCR (recognised as the reference method for sensitivity and specificity[3]) with the flexibility of compact, connected devices. The result is a network of distributed 'mini-laboratories' allowing reports within a couple of hours, revolutionising the management of diseases such as influenza A/B, RSV, SARS-CoV-2 and their possible co-infections. In the following paragraphs, we will analyse in detail how this technology works, what advantages it offers over rapid antigenic tests and what is the operational impact in clinical settings.
1. Technological structure and decentralised model
Decentralised Real-time PCR solutions are based on portable and networked instrumentation combined with cloud software for data control and analysis. A representative example is Ulysses Biomed's Hyris platform, consisting of the bCUBE™ device and the bAPP™ cloud application. bCUBE™ is a miniaturised thermal cycler, about the size of the palm of one hand (10x10x12 cm, ~1 kg), but capable of performing gene amplifications with laboratory-grade performance [4]. Despite its small size, this device supports rapid and controlled thermal cycling and can integrate up to 4 optical channels, allowing multiplex analysis with a broad spectrum (e.g. detecting multiple viruses simultaneously). Samples are housed in 16- or 36-well disposable cartridges, and multiple bCUBE units can operate in parallel, linearly increasing throughput [5]. Installation is plug-and-play: all that is needed is a power supply and a connection (Ethernet or Wi-Fi) to start operating, with no need for complex calibrations or laboratory infrastructure [6]. The brain of the system resides in bAPP™, a cloud software platform that allows remote control of one or more devices and access to real-time data [7]. Through an intuitive web interface, the operator can initiate preset or customised PCR protocols, monitor live amplification curves and receive an automatic interpretation of the result from the system. This last aspect is crucial: the built-in machine learning/AI algorithms support the analysis of complex outputs, e.g. recognising the presence of multiple pathogens from multi-channel fluorescent curves [8]. In practice, the software can provide an immediate report (e.g. 'positive for Influenza A and RSV'), reducing reliance on highly specialised personnel to interpret the raw data. All results are stored in the cloud database and accessible in real time from any location: this enables innovative operating models, such as remote supervision from a central laboratory or integration with hospital information systems (LIS/LIMS) for unified reporting. The model that emerges is that of distributed 'cloud-based' diagnostics: compact molecular devices located throughout the territory or in departments, orchestrated by a central intelligent platform.
2. Diseases covered and detectable co-infections
Influenza syndrome and other acute respiratory infections can be caused by a multiplicity of viruses and sometimes by virus-virus or virus-bacteria co-infections. Clinical symptoms (fever, cough, dyspnoea, sore throat, etc.) often overlap, making it difficult to distinguish the aetiology without laboratory testing. Multiplex PCR panels were created to address precisely this challenge: with a single nasopharyngeal swab, a broad spectrum of respiratory pathogens can be tested simultaneously. The decentralised platforms available today typically cover influenza A and B viruses, RSV (respiratory syncytial virus), SARS-CoV-2 and sometimes other common viruses (e.g. adenovirus, rhinovirus/enterovirus, metapneumovirus) depending on the kit used. Some syndromic molecular panels also include targets for atypical bacterial pathogens (e.g. Mycoplasma pneumoniae or Bordetella pertussis), or antibiotic resistance genes, providing a complete diagnostic picture in the case of pneumonia. Identifying a bacterial co-infection in a patient with influenza, for example, may direct towards a combination antiviral + antibiotic therapy, while confirming that a clinical picture is due solely to viruses may save antibiotics. Recent studies show that with the advent of molecular multiplex testing the detection of viral/bacterial co-infections has increased significantly, especially in community pneumonias [9]. This has important clinical implications: recognising a co-infection early on allows the patient to be managed more appropriately (e.g. by isolating the patient to prevent viral transmission, but also by administering targeted antibiotics for the identified bacterial pathogen early on).
In settings such as paediatric emergency rooms, where RSV and influenza circulate together during the winter season, having a rapid multiplex test makes it possible to detect double infections (e.g. influenza + streptococcus) that would otherwise be easily missed with single tests. Even during the COVID-19 pandemic, the simultaneous presence of SARS-CoV-2 and other respiratory pathogens (influenza in primis) was documented; this is why today many 'respiratory' multiplex tests include both influenza viruses and the SARS-CoV-2 coronavirus in a single panel, facilitating differential diagnosis and the possible detection of viral co-infections.
3. Limitations of rapid antigenic approaches
Before the widespread use of rapid PCR, decentralised diagnostics of respiratory infections were mainly based on rapid antigenic tests (e.g. immunochromatographic tests for influenza or RSV, and more recently antigenic tests for COVID-19). These assays offer results in 10-15 minutes and are simple to perform, but have important limitations in sensitivity. For example, Rapid Influenza Diagnostic Tests (RIDTs), while useful for immediate screening, may miss a significant proportion of positive cases. According to the CDC, influenza antigen tests should be interpreted with caution: sensitivity compared to PCR is sub-optimal, with frequent false negatives especially during periods of high viral circulation [10] [11]. In other words, a negative antigenic result does not really exclude active influenza infection, and in the presence of strong clinical suspicion the CDC still recommends considering antiviral therapy or confirming with a molecular test [12]. Similar limitations apply to RSV rapid tests and to many combined antigenic tests. The specificity of antigenic tests is generally good, but false positives may occur (especially for influenza B in some kits and in low disease prevalence) [13].
In summary, the antigenic approach suffers from a trade-off between speed and accuracy: it provides a fast response but risks underestimating cases, especially in the early stages of infection (when the viral load is at the limit of antigenic detectability) or for some viruses in particular. In addition, antigenic tests typically detect one pathogen at a time; there are combination kits (e.g. triple COVID/Influenza/RSV tests), but the level of multiplexing is limited and in any case each of these components retains the sensitivity constraints of the single antigen [14]. Multiplex PCR, on the other hand, offers both high sensitivity and the expansion of the number of targets that can be detected in parallel, combining the best of both worlds: speed and diagnostic reliability.
4. The added value of multiplex real-time PCR
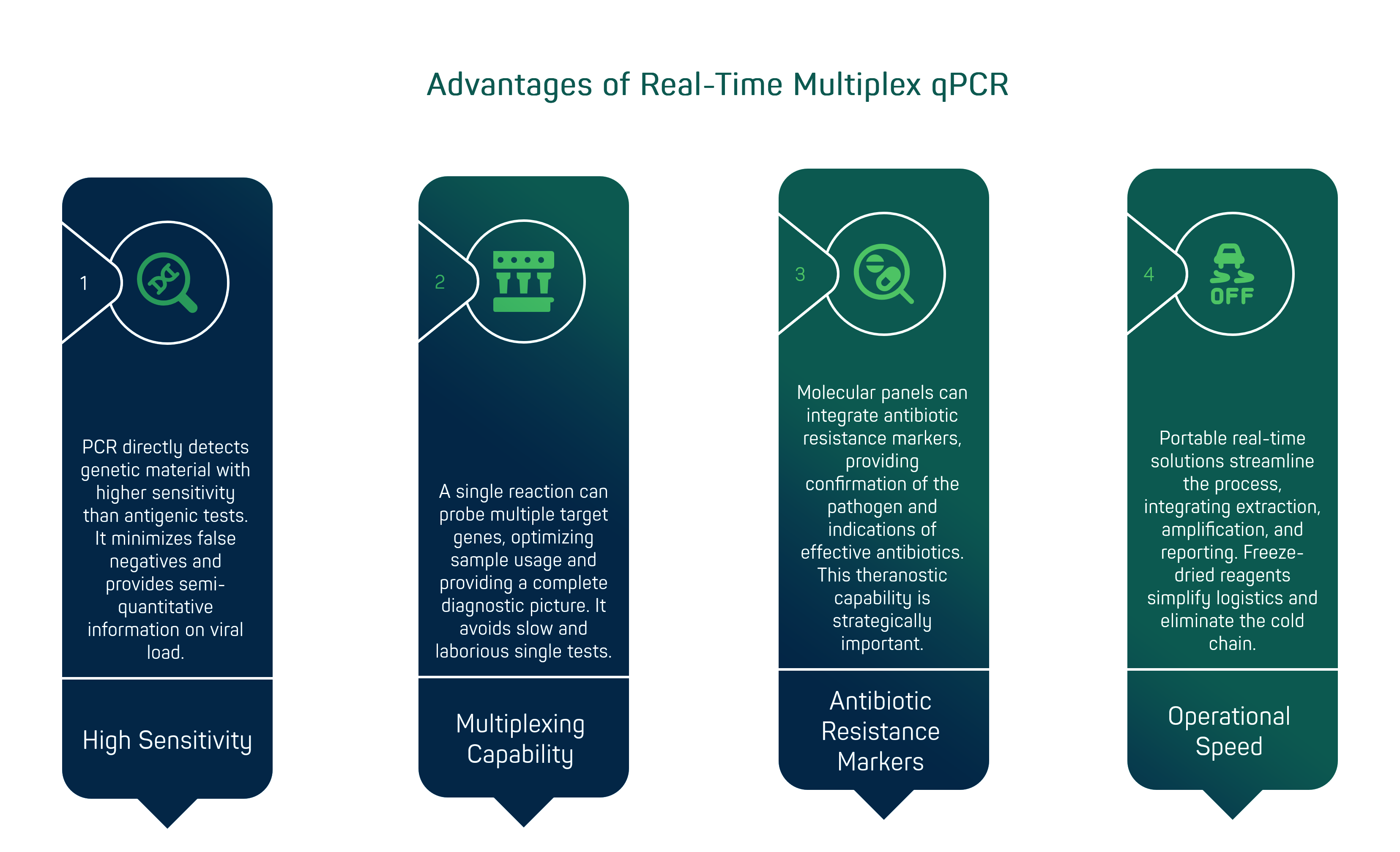
Real-time multiplex qPCR has several key advantages that make it a reference technology for advanced infectious diagnostics. Firstly, PCR directly detects the genetic material (RNA/DNA) of the pathogen with a much higher sensitivity than antigenic tests, especially in the early stages of infection or in the presence of low viral load. Nucleic acid amplification methods such as RT-PCR are now known to be the gold standard for confirming respiratory viral infections [3]. This results in an ability to identify virtually all infected cases, minimising false negatives. In addition, quantitative reading (Ct values, cycle threshold) provides semi-quantitative information on viral load, potentially useful for assessing the stage of infection or response to treatment.
The second advantage is inherent in the term multiplex: a single reaction can probe multiple target genes. For example, a multiplex respiratory panel can include primers and probes for influenza A (M gene), influenza B, RSV, SARS-CoV-2 (N gene and ORF1ab), plus possibly internal controls and bacterial targets. This approach optimises the sample - one sample for many tests - and offers a complete diagnostic picture with one examination. In clinical situations where symptoms are non-specific (cough, fever) but the list of possible aetiological agents is long, multiplex avoids the slow and laborious sequence of single tests (first influenza, if negative then RSV, if negative then... and so on).
A further added value is the possibility of integrating antibiotic resistance markers or genes characteristic of particular strains into molecular panels. For example, new-generation syndromic panels for hospital-acquired pneumonia combine the search for respiratory viruses and bacteria (S. pneumoniae, Legionella, etc.) with that of resistance genes (such as mecA for MRSA). In a single PCR session, the clinician obtains both confirmation of which pathogen is causing the infection and indications as to which antibiotics might (not) be effective. This theranostic (diagnosis + therapeutic indication) capability is of great strategic importance, especially in an era of worrying antimicrobial resistance.
From an operational point of view, speed remains the key issue: unlike traditional in-lab PCR protocols (which include extraction, amplification on standard thermal cyclers, and eventual report submission), portable real-time solutions integrate everything into a streamlined flow. Many devices accept the raw sample (nasopharyngeal aspirate, soil swab, etc.) without requiring manual RNA extraction, thanks to optimised chemical kits such as Ulysses Biomed's Sagitta line [15]. Freeze-dried reagents stable at room temperature simplify logistics by eliminating the cold chain, a critical feature for operating in remote environments or emerging countries [16]. In summary, the value of decentralised multiplex PCR lies in combining laboratory reliability with point-of-care readiness, enabling a new standard of rapid and comprehensive diagnosis directly at the patient's bedside.
5. bCUBE™ and bAPP™: a case of Italian innovation
As mentioned, the bCUBE™ + bAPP™ system from Ulysses Biomed represents one of the paradigms of this new decentralised diagnostics. It is worth exploring in more detail how these solutions are realised in practice. bCUBE™ is in essence a small portable molecular laboratory. Its technical features ensure that its analytical performance is comparable to that of benchtop thermal cyclers in a centralised laboratory [17]. This is confirmed by validation studies: in comparative tests, bCUBE demonstrated analytical accuracy equal to standard thermal cyclers and full compliance with international quality standards, being certified according to the European IVDR and equivalent North American regulations [18]. In terms of scalability, the modular architecture allows multiple bCUBE units to be added to the network: a hospital could deploy a device in each department (e.g. emergency room, intensive care unit, infectious ward) and have them operate simultaneously, connected to the same cloud platform.
For its part, bAPP™ acts as a central command console and repository. Through bAPP, a laboratory manager can remotely manage dozens of geographically dispersed analysers, see the live status of each test, and receive alerts upon completion. For example, a microbiology laboratory could instantly receive on its LIS system the result of a flu test done in an emergency room 2 km away, minutes after the patient was triaged. This makes it possible to centralise surveillance while decentralising execution: a model where the test takes place in the field, but the data immediately flows to a central hub to be aggregated, monitored and (if needed) notified to health authorities. Lorenzo Colombo, CTO of Ulisse Biomed, sums up the approach as follows: 'We have brought the quality of a centralised laboratory directly to the field. With bCUBE and bAPP, the laboratory follows the patient wherever they are needed, while the data automatically travels to the centre for real-time epidemiological analysis'. This inverted hub & spoke philosophy makes it possible to maintain high analytical standards while democratising access to advanced testing even in peripheral facilities or in situations where time is critical (outbreaks, emergencies, rural settings).
6. Use cases on the ward and in the territory
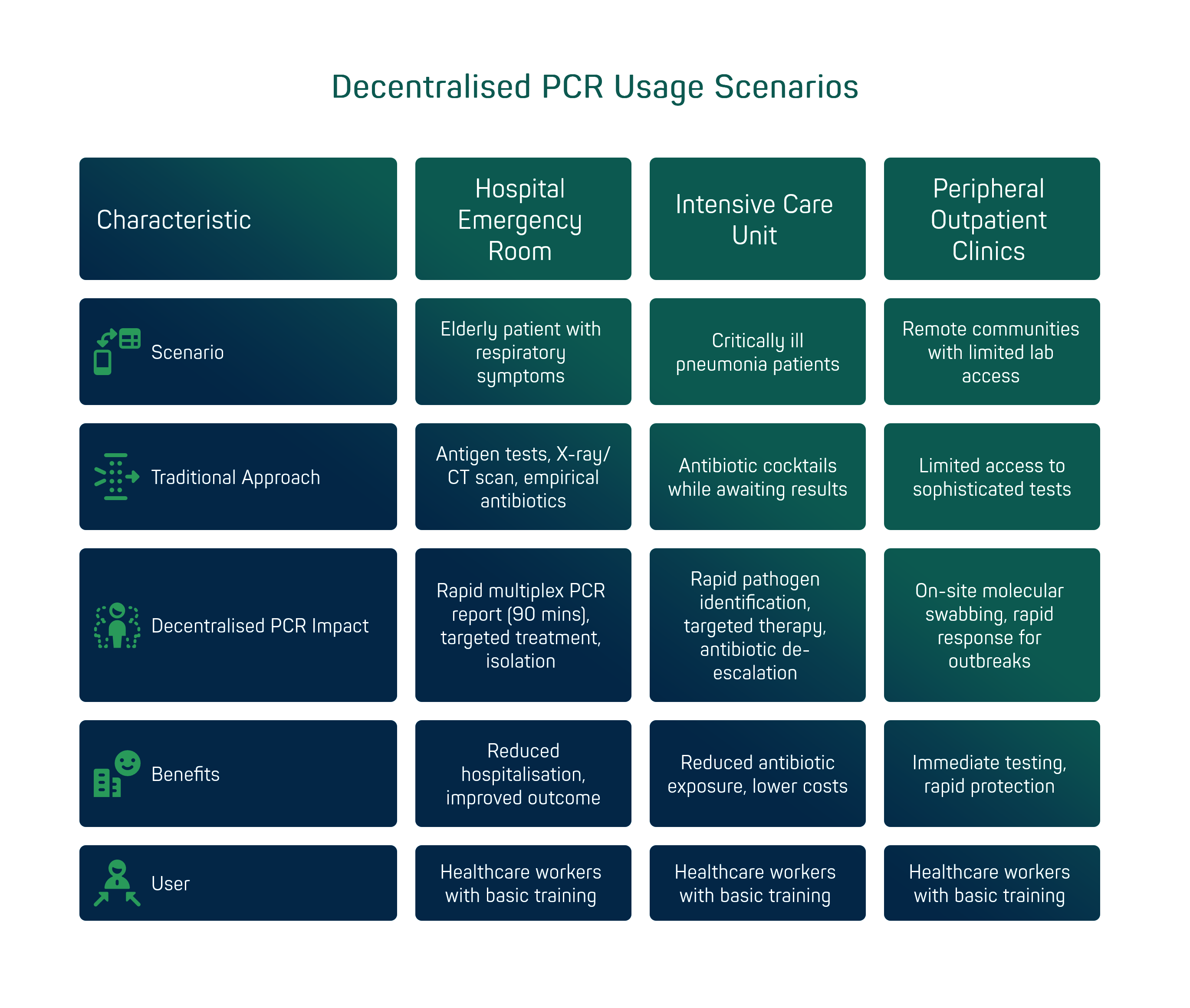
To understand the practical impact of decentralised PCR, it is useful to consider some typical usage scenarios. Hospital emergency room: an elderly patient arrives with a high fever, cough and breathlessness in the middle of winter. The symptoms could be due to influenza, COVID-19, bacterial pneumonia or a combination of these. In the traditional routine, one would perform an influenza and COVID antigen test (but have doubts about the reliability of a negative result) and in any case start an X-ray/CT scan for suspected pneumonia, administering empirical antibiotics in the meantime. With a point-of-care multiplex PCR system, on the other hand, the nasopharyngeal swab taken on arrival yields a complete report within 90 minutes: e.g. 'Influenza A and Streptococcus pneumoniae positive; RSV and SARS-CoV-2 negative'. This makes it possible in less than two hours to: isolate the patient in the influenza area (avoiding contagion in the ward), immediately start the specific antiviral (oseltamivir) for influenza A and at the same time administer the targeted antibiotic for pneumococcus (bacterial co-infection), perhaps avoiding other unnecessary broad-spectrum antibiotics. The result? Optimised management that can reduce hospitalisation time and markedly improve clinical outcome [1].
Intensive care unit: critically ill patients with severe pneumonia often receive antibiotic cocktails while waiting to clarify whether the origin is bacterial, viral or both. The use of extended PCR panels (such as those for nosocomial pneumonia) directly on the ward allows identification of the pathogens involved and resistance genes within a few hours. For example, detecting Staphylococcus aureus with a negative mecA gene could cause unnecessary anti-MRSA therapy to be suspended, or identifying a virus (metapneumovirus) behind an atypical respiratory picture would lead to immediate antibiotic de-escalation. These are decisions that traditionally would have waited 2-3 days (cultures, centralised results), whereas with decentralised diagnostics they take place within the same day, easing patient exposure to unnecessary antibiotics and reducing hospital costs.
Peripheral outpatient clinics and community clinics: in remote communities or simply outside large urban centres, access to sophisticated laboratory tests is often limited. A portable system such as bCUBE can be installed in a small district laboratory or even a clinical pharmacy, enabling services such as molecular swabbing for influenza and COVID with on-site response. This has been particularly useful during pandemic peaks, but it remains a valuable model beyond the pandemic: think of RSAs (residences for the elderly) where a cluster of influenza/RSV can have serious consequences - being able to immediately test all symptomatic patients on-site enables isolation of positives and rapid protection of other residents.
It should be emphasised that the user-friendliness of these solutions broadens their application contexts: healthcare workers with basic training (e.g. nurses, technicians) can perform tests thanks to guided interfaces and pre-assembled kits. This is crucial in environments such as field clinics, border stations or low-staffed wards: the technology is designed to fit seamlessly into the workflow.
7. Clinical and operational impact
The introduction of decentralised multiplex PCR in clinical settings has shown measurable tangible benefits. The first indicator is the dramatic reduction in reporting time (TAT). Meta-analyses of hospital studies indicate that the use of rapid sample-to-answer tests reduces turnaround time by approximately 24 hours compared to traditional PCR tests sent to the laboratory [1]. In practice, a result that with the usual route would arrive the next day (on average >24h), with point-of-care PCR is obtained in less than 2 hours after the sample is taken. This one-day delta is crucial in changing clinical outcomes. For example, one study found a reduction in average hospital stay of ~0.8 days for patients managed with rapid molecular diagnostics compared to those with standard procedures [1]. Discharging a patient almost a day earlier means freeing up beds, reducing the risk of additional nosocomial infections and cutting hospital costs.
Another significant impact is on therapeutic appropriateness. In the case of influenza, having a real-time result increases the likelihood that positive patients receive the appropriate antiviral therapy (e.g. oseltamivir) immediately by 25-30% [19]. At the same time, an increase of up to 55% in the timely adoption of isolation and infection control measures (single rooms, influenza cohort) has been observed for rapidly diagnosed patients [20]. This helps to contain the nosocomial spread of viruses and protect patients and caregivers.
On the antibiotic stewardship side, data suggest that the availability of rapid virological diagnosis leads to a reduction of empirical antibiotic use in patients with confirmed viral infections. In general, identifying a virus as the cause of respiratory symptoms allows clinicians to suspend or avoid antibiotics in those cases, focusing them only on patients with evidence of bacterial infection. Antimicrobial stewardship guidelines emphasise that rapid diagnostic tests should be integrated into stewardship programmes to reduce inappropriate antibiotic use [21]. Although studies in adults have yielded sometimes mixed results, several studies show (albeit moderate) reductions in antibiotic use and duration of therapy when viral results are communicated rapidly to the physician [22]. A practical example: in one hospital, switching from a traditional PCR panel with TAT ~27 hours to a rapid 1-2 hour panel showed a significant decrease in empirical antibiotic prescriptions in the first 24 hours of admission [23]. This indicates that physicians, having a rapid molecular test, are more likely to wait for the outcome to direct therapy instead of starting 'in the dark' with broad antibacterial coverage.
A further operational benefit is the reduction of additional diagnostic investigations. Knowing immediately that a patient has, for example, influenza A, can avoid a series of other tests (X-rays, repeated inflammatory markers, consultations) that are often requested in an attempt to clarify an uncertain diagnosis. A paediatric study showed that children who tested positive for influenza by rapid testing underwent fewer invasive and laboratory tests than those with a negative or uncertain diagnosis [24], an indication that a clear molecular diagnosis early in the course can streamline the clinical workup.
From an epidemiological and public health perspective, decentralised diagnostics finally offers an active surveillance advantage. By collecting results in the cloud in real time from various locations, an early warning system for outbreaks can be created: if an unusual increase in RSV-positive cases appears in various peripheral emergency rooms, the central laboratory or authorities can learn about it immediately and react (e.g. alert paediatric departments, check vaccination coverage, etc.). This concept of a distributed and interconnected diagnostic network turns every point of care into an epidemiological sentinel, without waiting for the established reporting times of centralised laboratories.
In summary, data and field experience confirm that decentralised Real-Time PCR is not just a technological fad, but results in better outcomes: faster diagnosis -> more targeted therapies -> shorter hospital stays -> less costs and less resistance. A win-win for both patients and the healthcare system.
"The widespread implementation of rapid and decentralised multiplex PCR tests raises the level of clinical response. In high-flow settings such as emergency rooms, going from a report in 24-48 hours to a report in 90-120 minutes radically changes decision-making: it means being able to isolate an infected patient immediately, start targeted therapies during triage and, most importantly, avoid unnecessary antibiotics when identifying a virus. From a technological point of view, we were able to bring the power of a centralised laboratory directly to the patient's bedside, with results also immediately available at the surveillance centre. This convergence of portable hardware and cloud software shortens the distance in healthcare: the right result, in the right place, at the right time."
— Lorenzo Colombo, CTO of Ulisse Biomed
Conclusions
Decentralised molecular diagnostics of respiratory diseases is now a concrete reality, resulting from the convergence of technological innovation and pressing clinical needs. We have seen how portable multiplex Real-Time PCR platforms, supported by cloud ecosystems and artificial intelligence algorithms, are redefining the time and manner of diagnosis in hospital and territorial settings. The benefits are multiple and synergistic: dramatic reduction of TAT, increased appropriateness in the use of antivirals and antibiotics, improved infection control measures and ultimately better clinical outcomes for patients. Moreover, this distributed model creates a real-time data network that can enhance epidemiological surveillance and response to emerging outbreaks, bridging the gap between territory and centre.
From a strategic point of view, investing in the decentralisation of diagnostics means making the healthcare system more responsive and resilient. An emergency room equipped with point-of-care PCR can effectively deal with both common seasonal flu and new pandemic threats, without waiting for external confirmation. At the same time, the centralisation of data via cloud platforms allows laboratories and health authorities to maintain quality control and overview, ensuring that this democratisation of diagnostics takes place in a safe and harmonised manner.
Of course, there are still challenges and room for improvement: from the still high cost per test compared to antigenic tests, to the need to train personnel in the proper use of these technologies, to full integration into existing clinical and organisational pathways. But the trend is set: as market analyses confirm, the decentralised molecular diagnostics segment (and portable PCR in particular) is growing at double-digit rates and is set to become a standard of care within the next decade. Authoritative organisations and scientific literature are beginning to openly acknowledge that 'routine mPOCT should replace laboratory-based diagnostics' at least in certain contexts and seasons.
In conclusion, decentralised Real-Time PCR for respiratory infections is no longer just a futuristic vision, but a current solution that is revolutionising rapid diagnostics. Hospital laboratories, clinics and emergency rooms now have the opportunity to embrace this paradigm shift, with proven clinical and economic benefits. The challenge now is to scale up these innovations, integrate them into standard protocols and ensure their economic sustainability, so that every patient - wherever they are - can get the right diagnosis at the right time.
Sources and bibliography
[1] [19] [20] Routine Rapid Multiplex PCR for Respiratory Viruses Improves Inpatient Outcomes - Pulmonology Advisor
[2] [10] [11] [12] [13] Rapid Influenza Diagnostic Tests | Influenza (Flu) | CDC
https://www.cdc.gov/flu/hcp/testing-methods/clinician_guidance_ridt.html
[3] [21] [22] [28] Multiplex Respiratory Virus Testing for Antimicrobial Stewardship: A Prospective Assessment of Antimicrobial Use and Clinical Outcomes Among Hospitalized Adults - PMC
https://pmc.ncbi.nlm.nih.gov/articles/PMC5853820/
[4] [5] [6] [7] [8] [15] [16] [17] [18] ID15 - The Ulisse Biomed Ecosystem: the Hyris System™ modular diagnostic platform
[9] Influenza and Bacterial Coinfection in Adults With Community ...
https://academic.oup.com/ofid/article/7/3/ofaa066/5760732
[14] Key Insights into Respiratory Virus Testing: Sensitivity and Clinical ...
https://www.mdpi.com/2076-2607/13/1/63
[23] Impact of multiplex molecular assay turn-around-time on antibiotic ...
https://www.sciencedirect.com/science/article/abs/pii/S138665321830283X
[24] Rapid Influenza Diagnosis in Children With Serious Bacterial ...
https://onlinelibrary.wiley.com/doi/10.1111/apa.70174
[25] Clinical impact of a routine, molecular, point-of-care, test-and-treat strategy for influenza in adults admitted to hospital (FluPOC): a multicentre, open-label, randomised controlled trial - PubMed
https://pubmed.ncbi.nlm.nih.gov/33285143/
[26] [27] Rapid point of care diagnostic tests for viral and bacterial respiratory tract infections--needs, advances, and future prospects - PubMed